Abstract
CRISPR systems are widespread in the prokaryotic world, providing adaptive immunity against mobile genetic elements1,2. Type III CRISPR systems, with the signature gene cas10, use CRISPR RNA to detect non-self RNA, activating the enzymatic Cas10 subunit to defend the cell against mobile genetic elements either directly, via the integral histidine–aspartate (HD) nuclease domain3,4,5 or indirectly, via synthesis of cyclic oligoadenylate second messengers to activate diverse ancillary effectors6,7,8,9. A subset of type III CRISPR systems encode an uncharacterized CorA-family membrane protein and an associated NrN family phosphodiesterase that are predicted to function in antiviral defence. Here we demonstrate that the CorA-associated type III-B (Cmr) CRISPR system from Bacteroides fragilis provides immunity against mobile genetic elements when expressed in Escherichia coli. However, B. fragilis Cmr does not synthesize cyclic oligoadenylate species on activation, instead generating S-adenosyl methionine (SAM)-AMP (SAM is also known as AdoMet) by conjugating ATP to SAM via a phosphodiester bond. Once synthesized, SAM-AMP binds to the CorA effector, presumably leading to cell dormancy or death by disruption of the membrane integrity. SAM-AMP is degraded by CRISPR-associated phosphodiesterases or a SAM-AMP lyase, potentially providing an ‘off switch’ analogous to cyclic oligoadenylate-specific ring nucleases10. SAM-AMP thus represents a new class of second messenger for antiviral signalling, which may function in different roles in diverse cellular contexts.
Main
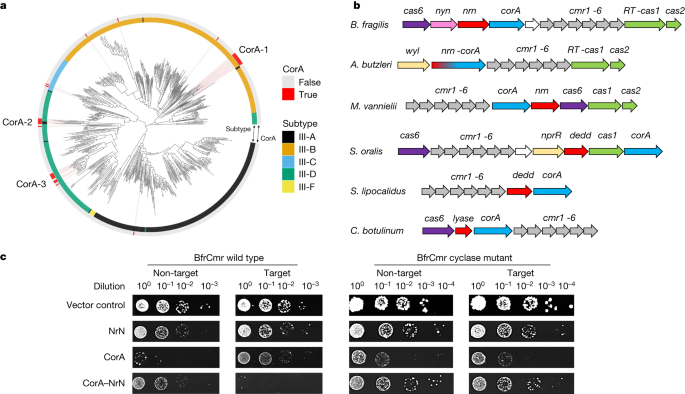
Bacteroides spp. are Gram-negative, anaerobic bacteria that constitute a significant portion of the human gut microbiome11. The Bacteroidales are host to the most widespread and abundant phage found in the human digestive system, CrAssphage12. B. fragilis is an opportunistic pathogen, and is responsible for more than 70% of Bacteroides infections13. Bioinformatic analyses have revealed the presence of three CRISPR types—I-B, II-C and III-B—in B. fragilis strains, with the type III-B system being the most common14. Sequence analysis shows that B. fragilis Cas10, the main enzymatic subunit of type III effectors, lacks an HD nuclease domain but has an intact cyclase domain, similar to the Vibrio metoecus Cas1015. This suggests that the system functions via cyclic oligoadenylate (cOA) signalling to associated ancillary effectors. In B. fragilis and more generally in the Cytophaga–Bacteroides–Flavobacterium bacterial phylum these type III CRISPR systems are strongly associated with an uncharacterized gene encoding a divergent member of the CorA-family of divalent cation channel proteins16,17 (Fig. 1a). The CRISPR-associated CorA proteins have not been studied biochemically but are predicted to consist of a C-terminal membrane spanning helical domain fused to a larger N-terminal domain with a unique fold. To investigate this further, we first generated a phylogenetic tree of Cas10 proteins and identified those associated with a gene encoding the CorA protein. Three phylogenetically distinct clusters of CorA-associated type III CRISPR systems were apparent, with the largest (CorA-1) being associated with type III-B systems (Fig. 1a).
a, A phylogenetic tree of Cas10 proteins from type III CRISPR systems of complete bacterial and archaeal genomes, colour coded by subtype42. Red bars on the outer ring indicate systems associated with a CorA-family effector protein. There are three main clusters of CorA-associated Cas10s, labelled CorA-1, CorA-2 and CorA-3. b, Genome context and effectors of selected type III-B CRISPR systems with a corA gene (cluster CorA-1: B. fragilis, Aliarcobacter butzleri, Methanococcus vannielii, Streptococcus oralis, Snytrophothermus lipocalidus, Clostridium botulinum). The type III-B cas genes cmr1–6 are shown in grey, with cas6 in purple and the adaptation genes cas1 (or a gene encoding a fused reverse transcriptase–Cas1 protein) and cas2 in green. The putative membrane channel protein is encoded by the corA gene (blue), which is adjacent to or fused with the genes encoding the PDEs NrN or DEDD (red). In C. botulinum, the PDE is replaced with a predicted SAM lyase. The wyl and nprR genes encode predicted transcriptional regulators. c, Plasmid challenge assay. E. coli BL21 Star cells expressing B. fragilis Cmr (wild-type or cyclase-defective variant) programmed with target (tetR) or non-target (pUC19) CRISPR RNA (crRNA) species were transformed with a pRAT plasmid that expressed the NrN and/or CorA proteins and carried a tetracycline resistance gene. Resistance was observed only when a targeting crRNA, active cyclase and both effector proteins were all present. Raw data are presented in Supplementary Data Fig. 1.
The genomic context of CorA-containing type III CRISPR loci from cluster CorA-1 (Fig. 1b) reveals that the corA gene is typically found next to a gene encoding a phosphodiesterase (PDE)—the DHH-family nuclease NrN in the case of B. fragilis and M. vanielii, and a DEDD-family nuclease in the case of S. oralis and S. lipocalidus. In the genome of A. butzleri and related species, the nrn and corA genes are fused, suggestive of a close functional relationship. The closest predicted structural matches for B. fragilis NrN are to the pGpG-specific PDE PggH from Vibrio cholerae, which has a role in the turnover of the cyclic nucleotide c-di-GMP18 and the GdpP PDE from Staphylococcus aureus, which degrades pApA molecules as a component of c-di-AMP signalling systems19. Analysis of the DEDD protein suggests structural matches to RNaseT20, oligoribonuclease21, and the mammalian REXO2 protein, which degrades linear RNA and DNA dinucleotides22. Thus, the CRISPR-associated NrN and DEDD proteins appear to be homologous to protein families that degrade small RNA and DNA species. Of note, in some CorA-containing type III systems including Clostridium botulinum, the PDE is replaced by a protein that is predicted to resemble a family of phage SAM lyase enzymes involved in evasion of host immune systems23,24 (Fig. 1b).
B. fragilis Cmr is active in vivo
To investigate the activity of the B. fragilis type III CRISPR system, two plasmids were constructed. Plasmid pBfrCmr1-6, built using Gibson assembly25, expresses synthetic versions of the codon-optimized genes cmr1–6 and plasmid pBfrCRISPR encodes Cas6 and a mini-CRISPR array (Extended Data Fig. 1a). We expressed the complex in E. coli with a targeting (pCRISPR-Tet) or non-targeting (pCRISPR-pUC) crRNA and challenged cells by transformation with a pRAT-Duet plasmid expressing one or both of the CorA and NrN effector proteins (Fig. 1c). The pRAT-Duet plasmid also contains the tetR gene for activation of B. fragilis Cmr carrying the targeting crRNA. Cells were transformed with the pRAT-Duet vectors and grown in the presence of tetracycline to select for transformants. We included vectors expressing wild-type and cyclase-defective (Cas10 D328A/D329A variant) Cmr for comparison. We previously used this experimental design to investigate the V. metoecus Cmr system15. In conditions in which the Cmr system was activated and had the required ancillary effector proteins, lower numbers of colony-forming units were expected. The vector control (no effectors) served as a baseline for transformation. When only the NrN effector was present, no reduction in colonies was observed, suggesting no active targeting. When only the CorA protein was expressed, fewer colonies were observed in both target and non-target conditions, for both the wild-type and cyclase-deficient mutant (Δcyclase) Cmr, suggesting some toxicity of the CorA protein. When both the CorA and NrN effectors were expressed, immunity—indicated by a markedly reduced number of colonies—was observed only for the wild-type Cmr system with tetR targeting. Immunity was lost when wild-type NrN was substituted with a variant mutated in the DHH active site motif (D85A/H86A/H87A), or when the CorA protein was truncated to remove the transmembrane domain (Extended Data Fig. 1b).
These data suggest that the B. fragilis Cmr system is functional in E. coli and requires the activity of the Cas10 cyclase domain and the presence of both effector proteins. The toxicity of CorA appears to be reduced by the presence of the NrN protein, regardless of activation of the type III system, suggestive of a strong functional link. Intriguingly, the type III-B complex from Mycobacterium tuberculosis, which synthesizes cyclic oligoadenylate 3–6 (cA3–6) in vitro26, did not provide immunity when combined with the NrN and CorA effectors, hinting at a non-canonical activation mechanism (Extended Data Fig. 1c). The strict requirement for the SAM-AMP degrading NrN protein in addition to CorA for plasmid immunity is an unusual aspect of the system and is discussed further below.
RNA processing and degradation
Co-transformation of the expression plasmids into E. coli strain BL21 (DE3) enabled the expression of the B. fragilis Cmr effector and purification by immobilized metal-affinity and size-exclusion chromatography (Extended Data Fig. 2a). We also purified B. fragilis Cas6 individually using the same chromatography steps. We first confirmed that Cas6 processed crRNA in the expected manner. The recombinant Cas6 enzyme cleaved synthetic fluorescein (FAM)-labelled crRNA at the base of a predicted hairpin with a 2-bp stem, reminiscent of Methanococcus maripaludis Cas6b27. This generates a canonical 8-nucleotide (nt) 5′ handle, (Extended Data Fig. 2b). Cleavage of an in vitro transcript comprising 2 repeats flanking one spacer generated the expected set of reaction products, culminating in a processed crRNA of 72 nt (Extended Data Fig. 2c). To investigate the composition of the crRNA present in the effector complex purified from E. coli, we isolated and labelled the crRNA using γ-32P-ATP and polynucleotide kinase. This revealed 3 major crRNA species differing in length by 6 nt (Extended Data Fig. 3a,b). These products correspond to 3′ end trimming of the crRNA to remove the repeat-derived sequence and probably reflect effector complexes that differ in the number of Cas7 subunits and thus length of backbone, as has been seen for other type III systems (reviewed in ref. 28).
Type III CRISPR systems also cleave bound target RNA using the Cas7 subunit, either for direct defence against mobile genetic elements29 or for regulatory purposes8. We proceeded to test for cleavage of target RNA bound to the crRNA in the effector. The 5′-end-labelled target RNA was cleaved at 4 positions with 6-nt spacing, corresponding to the placement of the Cas7 active sites in the backbone29. Cleavage was extremely rapid and was essentially complete after 2 min, the first time point (Extended Data Fig. 3c). As these sites interconvert and site 1 is furthest from the 5′ label, this cleavage was only observed for the Cmr4 or Cas7 D27A variant, which cleaves target RNA more slowly (Extended Data Fig. 3d). We also observed cleavage of target RNA at the boundary of the crRNA–target RNA duplex. This activity, which has not been observed for other type III systems, appeared to be due to the Cmr4 subunit, as it was not observed for the D27A variant. As target RNA cleavage has been shown to correlate with the deactivation of the Cas10 subunit8,30, this suggests that the Cmr complex remains active for a very short time after target RNA binding. This groups B. fragilis Cmr together with the type III effectors from Streptococcus thermophilus and Thermotoga maritima, which cleave target RNA rapidly5,30. By contrast, the type III systems from S. solfataricus and V. metoecus have much slower RNA cleavage kinetics8,15. In the absence of structural data to define the number and positions of Cas7 subunits in the complex, we could not analyse the cleavage pattern further with any degree of certainty.
Identification of the signalling molecule
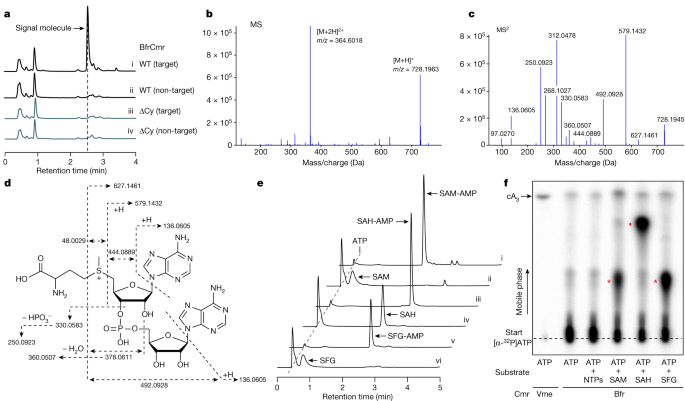
As B. fragilis Cmr lacks a HD nuclease domain in the Cas10 subunit, immune function would be expected to be mediated by the cyclase domain via the generation of nucleotide second messengers. However, although the system provided cyclase-dependent immunity in vivo, activation of the wild-type Cmr in vitro resulted in very low yields of any observable product when incubated with ATP, in contrast to the Cmr complex from V. metoecus, which synthesizes cA315. This hinted at the possibility that a vital component was missing in the in vitro assays. Accordingly, we activated B. fragilis Cmr in E. coli using a plasmid to express target RNA and then processed cell lysates to allow isolation of nucleotide products. These were purified and analysed by high-performance liquid chromatography (HPLC). A prominent peak was observed following HPLC of extracts with activated Cmr, which was absent in the absence of target RNA or when the cyclase activity was knocked out by mutagenesis (ΔCy) (Fig. 2a). Mass spectrometry yielded a m/z value of 728.196 for the positive ion; to our knowledge, this m/z value did not correspond with any known cyclic nucleotide or indeed any other previously characterized metabolite (Fig. 2b). To identify the product, we fragmented the purified molecule using tandem mass spectrometry (MS/MS). This enabled the identification of fragments characteristic of AMP and methionine (Fig. 2c). Further examination suggested that the molecule under study was SAM that was adenylated on the ribose moiety (Fig. 2d), a molecule that we hereafter refer to as SAM-AMP. To our knowledge, SAM-AMP has not previously been described in the literature—from either chemical or enzymatic synthesis perspectives—suggesting that it is a previously undiscovered class of signalling molecule.
a, HPLC analysis of E.coli extracts expressing the wild-type (WT) or mutant (ΔCy) B. fragilis Cmr system with target or non-target crRNA. The putative signal molecule was only observed for the activated system (trace i). b, Characterization of the extracted signal molecule by liquid chromatography–mass spectrometry (LC–MS) in positive mode. [M + H]+ and [M + 2H]2+ represent two different ionization forms. c, MS/MS analysis of the signal molecule with m/z 728.1963. d, The proposed structure of the signalling molecule, whose fragmentation pattern is shown by dashed arrows. The MS/MS data cannot distinguish between 2′–5′ and 3′–5′ phosphodiester bonds. The 3′–5′ phosphodiester bonds are more likely and is shown here, but a 2′–5′ bond cannot be completely ruled out. e, HPLC analysis of compounds synthesized by the purified wild-type B. fragilis Cmr complex in vitro. Cmr synthesizes the signal molecule SAM-AMP from ATP and SAM (trace i). Cmr also accepts SAH and sinefungin (SFG) as substrates (traces iii and v, respectively). Traces ii, iv and vi are control reactions. f, TLC analysis of in vitro reaction products. SAM, SAH and sinefungin plus ATP yielded radioactive products (red stars) but ATP alone did not. cA3 generated by wild-type V. metoecus Cmr complex15 is shown for comparison. Uncropped HPLC and TLC data are presented in Supplementary Data Fig. 2.
To confirm that B. fragilis Cmr synthesized SAM-AMP, we reconstituted the reaction in vitro with ATP and SAM, analysing reaction products by HPLC and thin-layer chromatography (TLC) (Fig. 2e,f). We observed SAM-AMP production when SAM and ATP were present in vitro. S-adenosyl-l-homocysteine (SAH) and the SAM analogue sinefungin31, which differ at the sulfur centre, were also conjugated with ATP by Cmr (Fig. 2e,f and Extended Data Fig. 4c). No significant products were observed in the presence of ATP or all four ribonucleotides. The synthesis of SAM-AMP and SAH-AMP by B. fragilis Cmr were consistent with rapid, multiple-turnover kinetics that were essentially complete within the first 2 min of the reaction (Extended Data Fig. 4a,b). The observation of only SAM-AMP, and not SAH-AMP, in E. coli cell extracts is probably the result of the much higher concentration of SAM than SAH in E. coli32 (0.4 mM versus 1.3 µM). Overall, these data provide strong evidence that the B. fragilis Cmr system generates a previously undescribed conjugate of SAM and ATP, rather than cOA.
Since Cas10 family enzymes synthesize 3′–5′ phosphodiester bonds6,7, we considered it likely that SAM was fused to AMP at the 3′ position on the ribose ring, but the mass spectrometry data did not rule out a 2′–5′ phosphodiester bond. To address this, we incubated SAM-AMP and SAH-AMP with nuclease P1, which is specific for 3′–5′ phosphodiester bonds. Whereas SAH-AMP was completely degraded by nuclease P1, we observed only partial degradation of SAM-AMP (Extended Data Fig. 5). Thus, although we consider a 3′–5′ phosphodiester linkage to be likely, we cannot rule out a 2′–5′ linkage completely. Final confirmation of the linkage will require further analysis—for example, by NMR.
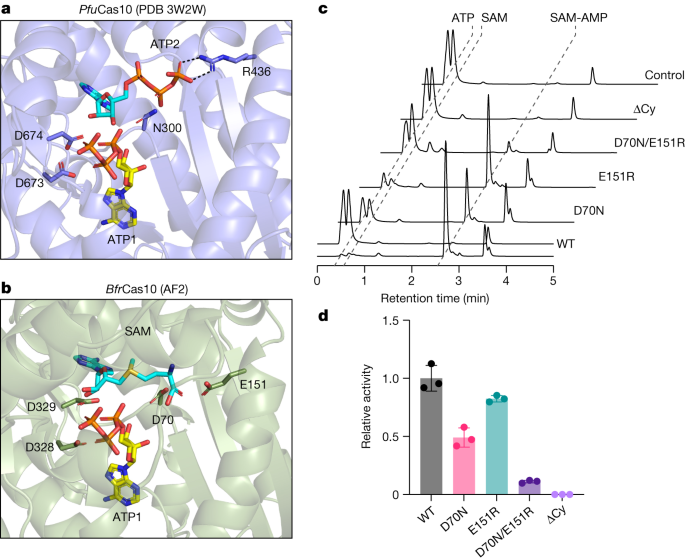
The crystal structure of Pyrococcus furiosus Cas10–Cas5 bound to two ATP molecules33 shows one ATP in the ‘donor’ ATP1 site next to the GGDD cyclase catalytic motif and another in the ‘acceptor’ site (Fig. 3a). For the enzymes that synthesize SAM-AMP, SAM must bind in the acceptor ATP2 binding site, next to ATP1 in the donor site34. This arrangement would allow nucleophilic attack from the 3′-hydroxyl group of SAM to the α-phosphate of ATP1, resulting in the formation of a 3′–5′ phosphodiester bond linking SAM with AMP, and the release of pyrophosphate. The reaction chemistry is essentially the same as the one that takes place in canonical type III CRISPR systems that synthesize cOA species34. The major difference is that the triphosphate of ATP2 in the acceptor site is replaced by the methionine moiety of SAM, resulting in a change in local charge of the ligand from net negative to net positive, raising the possibility that Cas10s binding SAM will have a less basic binding site in this area. Examination of sequence conservation in the Cas10s associated with a CorA-1 cluster (Extended Data Fig. 6) revealed the presence of two absolutely conserved acidic residues, D70 and E151. Modelling of the B. fragilis Cas10 structure places these two residues in the vicinity of the methionine moiety of SAM in the acceptor site (Fig. 3b). D70 occupies the position equivalent to N300 in P. furiosus Cas10, which neighbours the β-phosphate of the acceptor ATP2 ligand, while E151 is in a similar position to R436, which forms a bidentate hydrogen bond with the γ-phosphate (Fig. 3a). We created variants of Cas10 with D70N, E151R and D70N/E151R mutations, which were expressed and purified as for the wild-type protein (Extended Data Fig. 2a), and assessed them for their ability to synthesize nucleotide products (Fig. 3c). The E151R variant had a limited effect on SAM-AMP synthase activity, but the D70N variant was significantly compromised and the double mutant showed no detectable activity (Fig. 3c,d). Moreover, the double mutant displayed an enhanced pppApA synthase activity when compared with the wild-type enzyme, suggesting a partial reversion of the acceptor binding site to favour ATP over SAM (Extended Data Fig. 7). A deeper understanding of the reaction mechanism and substrate specificity of the SAM-AMP synthases will require structural data in the presence of ligands, and could also involve discrimination by the Cas5 subunit, which is in the vicinity of the ATP2 ligand.
a, The crystal structure of the P. furiosus (Pfu) Cas10 subunit with 2 bound ATP molecules33. Side chains for the two metal binding aspartate residues of the DD motif, together with residues N300 and R436 that interact with ATP2, are shown. b, Equivalent view of the AF2 model of the B. fragilis (Bfr) Cas10 structure with ATP1 from the P. furiosus Cas10 structure and ATP2 replaced by SAM. The precise conformation and position of SAM is unknown. The conserved acidic residues D70, E151, D328 and D329 are shown. c, In vitro SAM-AMP synthase activity of wild-type and variant B. fragilis Cmr, analysed by HPLC following incubation of 2 µM Cmr with 0.5 mM ATP and SAM for 30 min. Raw HPLC data are presented in Supplementary Data Fig. 3. d, Relative SAM-AMP synthase activity of Cmr variants. Three independent experiments; data are mean ± s.d., calculated using GraphPad Prism 9.
SAM-AMP signalling and turnover
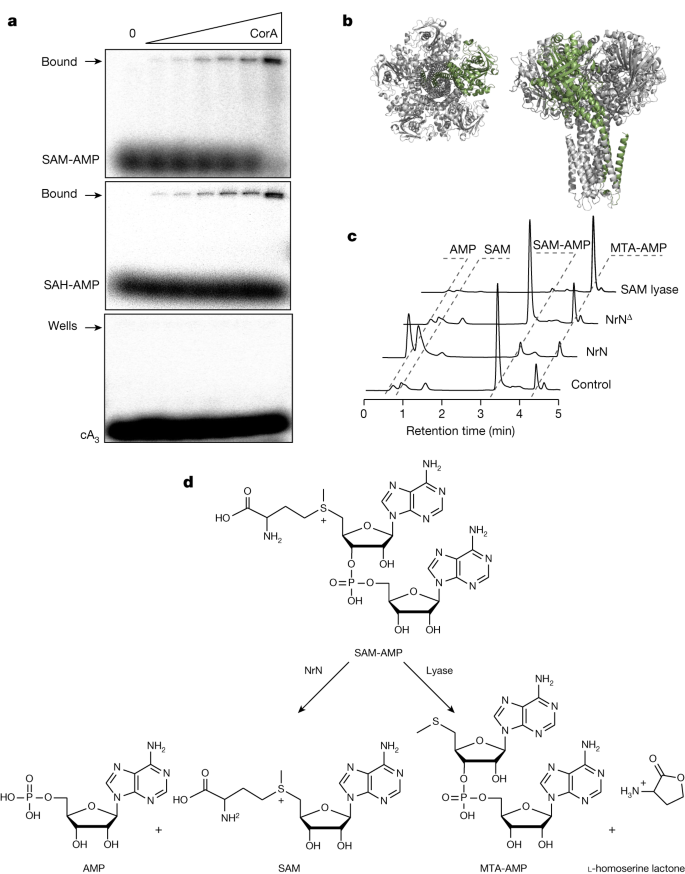
To test the suggestion that SAM-AMP is the activator of the CorA effector, we expressed B. fragilis CorA in E. coli and purified the protein to near homogeneity in the presence of detergent (Extended Data Fig. 8a). CorA was incubated with radiolabelled SAM-AMP, SAH-AMP or cA3 and then analysed by native gel electrophoresis. A clear shifted species, close to the wells of the gel, was observed to accumulate as the CorA protein was incubated at increasing concentrations with SAM-AMP or SAH-AMP (Fig. 4a). By contrast, cA3 was not shifted. These data support a model where CorA binds the SAM-AMP second messenger to provide immunity. To investigate this in more detail, we generated a model of the pentameric B. fragilis CorA structure (Fig. 4b and Extended Data Fig. 8) and mapped the positions of conserved residues in the CorA-1 clade identified from a multiple sequence alignment (Supplementary Data Fig. 10). A cluster of conserved residues at the interdomain interface hinted at a putative SAM-AMP-binding site. To test this, we created two site-directed variants of CorA by mutating two pairs of conserved residues (R152–R153 and D219–D220) in this cluster to alanine. The variant proteins were expressed similarly to the wild-type CorA, but no longer provided immunity in the plasmid challenge assay (Extended Data Fig. 8). Although these observations are consistent with a role in SAM-AMP binding, we cannot rule out the possibility that these mutations alter the quaternary structure of the protein. Although the mechanism of the CorA effector has not yet been determined, it most probably functions as a SAM-AMP-activated membrane channel, analogous to the Csx28 protein associated with Cas1335 and to a number of other predicted membrane proteins associated with type III CRISPR systems16
a, CorA binds SAM-AMP and SAH-AMP, but not cA3 (1 µM 32P-labelled ligand incubated with 0, 0.0625, 0.125, 0.35, 0.75, 1.5 and 3.3 µM CorA), illustrated by acrylamide gel electrophoresis and phosphorimaging. Uncropped gels are shown in Supplementary Data Fig. 4. b, Two views of the predicted structure of the pentameric CorA channel, with one subunit coloured green. c, NrN specifically degrades SAM-AMP to SAM and AMP. HPLC analysis of samples in which purified SAM-AMP was incubated with NrN and NrNΔ, an inactive variant with D85A/H86A/H87A mutations. C. botulinum lyase degrades SAM-AMP to generate MTA and l-homoserine lactone (not UV visible). Small amounts of MTA are present in the SAM-AMP sample purified from E. coli. Uncropped HPLC traces are available in Supplementary Data Fig. 4. d, Schematic representation of the reactions catalysed by NrN and SAM-AMP lyase.
As described previously, most type III systems with a CorA effector also encode a PDE of the NrN or DEDD family16. We therefore incubated the B. fragilis NrN protein with SAM-AMP and observed that it specifically degrades SAM-AMP (Fig. 4c), but not the linear dinucleotide pApA, or cOA molecules cA2–6 (Extended Data Fig. 9a). One possibility is that specialized NrN and DEDD-family PDEs represent a type of ‘off switch’ to reset the system, analogous to the ring nucleases that degrade cOA molecules in canonical type III CRISPR systems10. In the Clostridia, the NrN protein is replaced with a predicted SAM lyase (Fig. 1b), suggesting an alternative means to degrade the SAM-AMP signalling molecule. We tested this by cloning and expressing the SAM lyase from C. botulinum and measuring its ability to degrade SAM-AMP, observing efficient degradation of the molecule to 5′-methylthioadenosine (MTA) (Fig. 4c,d). The other product of a lyase reaction, l-homoserine lactone, is not detectable by UV. The C. botulinum lyase degrades SAM-AMP more efficiently than SAM (Extended Data Fig. 9b), consistent with a specialized role in defence.
Discussion
The polymerase active site of Cas10, the catalytic subunit of type III CRISPR systems, which consists of two DNA polymerase family B palm domains, is known to synthesize a range of cOA second messengers for antiviral defence. Here we have shown that some type III CRISPR systems signal via synthesis of SAM-AMP, a previously unknown molecule created by the adenylation of SAM (Fig. 5), which thus represents a novel nucleotide-based second messenger. In bacteria, the most recently discovered anti-phage signalling molecules include the cUMP and cCMP of the PYCSAR system36, a wide range of cyclic di- and tri-nucleotides of the CBASS system37 and the cOAs typically made by type III CRISPR systems6,7. Given the structural similarity between ATP and SAM, it is perhaps not surprising that SAM can substitute for ATP as an acceptor for a new 5′–3′ phosphodiester bond in the active site of nucleotide cyclases, following limited sequence divergence. Clearly this reaction reaches a natural end point as there is no possibility of cyclization or further polymerization. Judging by the distribution of CorA effectors, SAM-AMP signalling has a patchy but wide distribution in members of the bacteroidetes, firmicutes, δ-proteobacteria, ε-proteobacteria and euryarchaea. This is consistent with the high levels of defence system gain (by lateral gene transfer) and loss observed generally, and may be a reflection of the pressures exerted by viruses, driving diversity.
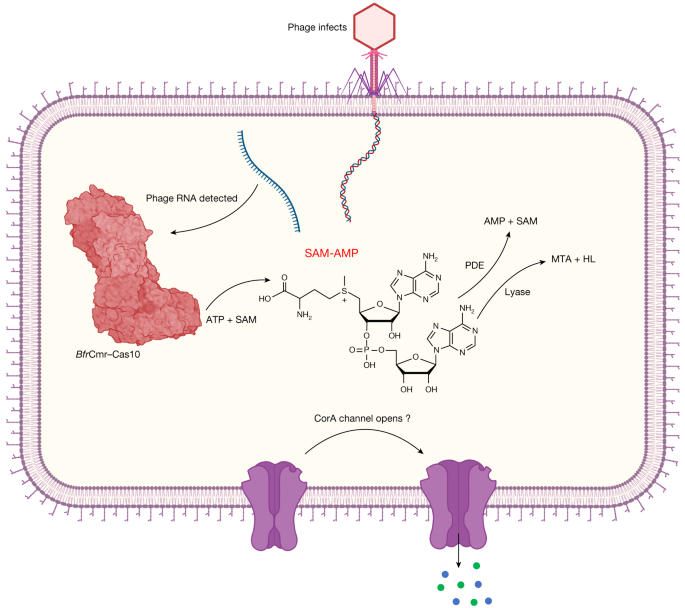
Transcription of the infecting phage genome activates the B. fragilis Cmr complex, resulting in synthesis of the SAM-AMP second messenger. SAM-AMP binds to the CorA membrane protein, resulting in the opening of a pore that disrupts the host membrane to combat infection. SAM-AMP is degraded by specialized PDE enzymes that hydrolyse the phosphodiester bond, generating AMP and SAM or lyases that target the methionine moiety, generating MTA and homoserine lactone (HL). These enzymes are likely to deactivate the signalling molecule to reset the system once phage have been eliminated. Created with BioRender.com.
CRISPR-associated CorA proteins are predicted to have a N-terminal soluble domain fused to a C-terminal transmembrane helical domain related to the CorA family of divalent cation transporters16. We postulate that binding of SAM-AMP to the cytoplasmic domain results in an opening of the transmembrane pore to effect immunity, but alternative mechanisms of membrane disruption have been observed for bacterial immune effectors38, so this is a priority for future studies. The CorA effectors seem to be obligately associated with degradative enzymes such as NrN in B. fragilis, sometimes even being fused16. The observation that SAM-AMP is easily purified from extracts of E. coli expressing the activated B. fragilis Cmr system suggests that SAM-AMP is not a substrate for the generalist ribonucleases present in bacteria, necessitating the addition of a specialized PDE such as NrN. In these respects, NrN is reminiscent of the ring nucleases (Crn1–3 and Csx3) that are frequently found associated with cOA generating CRISPR systems10. This suggests that it is beneficial to the cell to deplete the SAM-AMP signalling molecule, perhaps to avoid unnecessary cell death when phage infection has been cleared. In this regard, it is telling that the NrN PDE is sometimes replaced by a SAM-AMP lyase—an enzyme that degrades SAM-AMP using an entirely different mechanism23, yielding different products. SAM lyases are typically phage related genes and are thought to function by neutralizing DNA methylases in host restriction-modification systems23,24. In the context of CorA-family CRISPR systems, SAM-AMP lyases encoded by mobile genetic elements may also function as anti-CRISPRs, similarly to viral ring nucleases39.
Some important open questions remain. It is difficult to explain the toxicity of CorA when no SAM-AMP is synthesized and NrN is absent, as well as the observation that both CorA and NrN are required for immunity. These data suggest a close functional link between CorA and NrN, although we have detected no physical interaction between the two proteins in vitro. Rather than functioning in a manner analogous to ring nucleases, an alternative hypothesis is that NrN (or SAM-AMP lyase) is required to prevent de-sensitization of the CorA channel—a phenomenon observed for other pentameric ligand-gated ion channels when activator concentrations remain high40. Answers to these questions will probably require further analysis of the system in a cognate host at native expression levels, coupled with structure–function studies of the CorA channel.
Given the wide range of SAM and ATP analogues available, the discovery of an enzymatic route to synthesis of SAM-AMP opens the way to the generation of a new family of bioactive molecules. For example, there is considerable interest in the development of specific inhibitors of methyltransferases, a large family of enzymes (more than 300 methyltransferases are encoded in the human genome) involved in many key cellular reactions41. Depending on the specificity of the Cas10 enzyme, a range of SAM and ATP analogues could be provided as building blocks to make a diverse family of SAM-AMP analogues with altered properties. As we have seen, replacement of the methyl group on the sulfur atom of SAM with a proton (in SAH) or an amino group (sinefungin) still supports catalysis by B. fragilis Cas10. Many other modification sites are available on the parental molecules.
In conclusion, we report the discovery of SAM-AMP, which is synthesized from two of the most abundant molecules in the cell and functions as a second messenger of viral infection. This broadens the repertoire of type III CRISPR systems and may have implications for immune signalling more generally, as family B polymerases are a widespread and diverse superfamily found in all branches of life. The recent expansion of our knowledge of signalling molecules reflects the fact that Nature tends to use and repurpose such molecules in diverse cellular processes.
Methods
Cloning
Supplementary Table 1 shows the synthetic gene, DNA and RNA oligonucleotide sequences used in this study.
The synthetic genes encoding B.fragilis Cas6, CorA, NrN and C. botulinum SAM lyase purchased as g-blocks (IDT) were codon-optimized for expression in E. coli C43 (DE3) via the vector pEhisV5TEV, which encodes eight histidines, a V5 epitope tag and the cleavage site of Tobacco Etch Virus (TEV) protease43.
The pACE-based Cmr expression plasmid pBfrCmr1-6 was designed to contain six codon-optimized genes cmr1–6 (Twist Biosciences), with a polyhistidine tag on the N-terminus of Cmr3. We then divided pBfrCmr1-6 into five overlapping segments with similar length (designated as BfrCmr a, b, c, d and e). These segments were amplified by PCR using primers BfrCmrSG-f and BfrCmrSG-r (Supplementary Table 1) and then assembled into pBfrCmr1-6 using NEBuilder HiFi DNA Assembly Master Kit. The obtained plasmid pBfrCmr1-6 was verified by restriction digest and sequencing.
For the construction of the crRNA over-expression vector, the codon-optimized cas6 gene was synthesized as a g-Block (IDT) and inserted into the NdeI and XhoI restriction sites in MCS-2 of the vector pCDFDuet. The synthetic gene of CRISPR pre-array with two CRISPR repeats and two divergent BpiI sites between two repeats for a spacer sequence insertion was cloned into 5′ NcoI and 3′ SalI sites in MCS-1 of pCDFDuet containing cas6. Spacers targeting the tetracycline resistance gene tetR or lacZ were ligated into the BpiI sites of CRISPR pre-array to obtain the plasmid, designated as pCRISPR_Tet or pCRISPR_pUC. A CRISPR array, consisting of one spacer targeting the gene encoding late promoter activating protein (Lpa) of phage P1, flanked by two B. fragilis CRISPR repeat sequences, was assembled by annealing primers Bfr-rep-5p-T, Bfr-rep-5p-C, Bfr-rep-3p-T, Bfr-rep-3p-C, Bfr-sp-phageIPA-T and Bfr-sp-phageIPA-C (Supplementary Table 1). The array was then ligated into MCS-1 of pCDFDuet containing cas6 in MCS−2 to give pCRISPR_Lpa.
To construct pRAT-Duet-derived plasmids, for single effector expression, synthetic genes encoding CorA, NrN or their variants were inserted into NcoI and EcoRI sites in MCS-1 under control of the pBAD promoter. The nrn gene was cloned into the NdeI and XhoI site in MCS-2 of pRAT-Duet containing corA in MCS-1 for effector co-expression.
For mutagenesis, all variants were generated by PCR using Phusion enzyme (Thermo Scientific) with wild-type constructs as the templates in presence of two overlapping primers containing the target mutations. The correct variants were confirmed by sequencing.
Protein production and purification
B. fragilis NrN, Cas6 and C. botulinum SAM lyase were expressed and purified using standard procedures described previously, with removal of the N-terminal his-tags by TEV protease43. The Cmr complex with crRNA expressed from cells co-transformed with pACE-BfrCmr and pCRISPR_Lpa was purified using the same procedures. The purification of membrane protein BfrCorA was followed the similar procedures. In brief, after induction of CorA expression with 0.2 mM IPTG, the cells were harvested and lysed in the lysis buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 5% glycerol, 10 mM imidazole). The membrane fraction was isolated by ultra-centrifuge at 40,000 rpm at 4 °C for 2 h and then resuspended in the lysis buffer supplement with 1% n-dodecyl-β-d-maltopyranoside (Glycon Biochemical) and incubated at 4 °C for 1 h. The solubilised CorA was purified by nickel affinity chromatography and size-exclusion chromatography, where buffers contained 0.03% n-dodecyl-β-d-maltopyranoside. Subsequently, a series of variants were constructed and purified following the same procedure as the wild type.
Plasmid challenge assay
pBfrCmr1–6 and pCRISPR_Tet (or pCRISPR_pUC) were co-transformed into E. coli BL21 Star cells. Single colonies were picked for competent cell preparation into L-Broth (100 µg ml−1 ampicillin and 50 µg ml−1 spectinomycin) and cultivated at 37 °C overnight. Overnight culture was diluted 50-fold into 20 ml selective LB medium and grown at 37 °C with shaking at 220 rpm until the OD600 reached 0.4–0.5. Cell pellets were collected and then resuspended in an equal volume of pre-chilled competent cells solution (60 mM CaCl2, 25 mM MES, pH 5.8, 5 mM MgCl2, 5 mM MnCl2). Cells were incubated on ice for 1 h, centrifuged and the collected pellet was resuspended in 0.1 volumes of the same buffer containing 10% glycerol. Aliquots (100 µl) were flash frozen by liquid nitrogen and then stored at −80 °C. 100 ng pRAT or pRAT derived plasmids carrying the target gene were added to the competent cells, incubated on ice for 30 min and transformed by heat shock. Following addition of 0.5 ml LB medium, the transformation mixture was incubated at 37 °C for 2.5 h. 3 µl of a 10-fold serial dilution was applied in duplicate to LB agar plates (supplemented with 100 µg ml−1 ampicillin and 50 µg ml−1 spectinomycin) for the uninduced condition. The transformants were selected on LB agar containing 100 µg ml−1 ampicillin, 50 µg ml−1 spectinomycin and 12.5 µg ml−1 tetracycline. Addition of 0.2% (w/v) lactose and 0.2% (w/v) l-arabinose was used for full induction. Plates were incubated at 37 °C overnight. The experiment was performed as two independent experiments with two biological replicates and at least two technical replicates.
Synthesis of SAM-AMP and its analogues
For in vitro synthesis, 2 µM wild-type Cmr was incubated with ATP and SAM, SAH or sinefungin (0.1 mM each for radiolabelled products or 0.5 mM each for HPLC analysis) in reaction buffer (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, 1% glycerol and 5 mM MnCl2). The reaction was initiated by adding 5 µM target RNA–Lpa (using non-target RNA-pUC as negative control) and carried out at 37 °C for 1 h or the times indicated. 4 nM α32P-ATP as a tracer was added in each reaction to generate radiolabelled products, if needed for TLC analysis or electrophoretic mobility shift assay (EMSA).
For in vivo production, a single colony of E. coli BL21 Star cells transformed with the plasmids pBfrCmr1-6, pCRISPR_Tet (or pCRISPR_pUC) and pRAT-Duet was inoculated into 10 ml of L-broth with antibiotics (50 µg ml−1 ampicillin, 50 µg ml−1 spectinomycin and 12.5 µg ml−1 tetracycline) and grown overnight at 37 °C with shaking at 180 rpm. 20-fold overnight culture was diluted into 20 ml fresh L-broth with same antibiotics and then incubated at 37 °C. The culture was fully induced with 0.2% (w/v) d-lactose and 0.2% (w/v) l-arabinose after reaching an OD600 between 0.4 and 0.6. After overnight induction at 25 °C, the cell culture was mixed with 4 volumes of cold PBS and then centrifuged at 4,000 rpm for 10 min at 4 °C. Cell pellet was resuspended into 2 ml cold extraction solvent (acetonitrile:methanol:water, 2:2:1 by volume), vortexed for 30 s and stored at −20 °C until needed. The supernatant was obtained by centrifuging at 13,200 rpm for 10 min at 4 °C, followed by evaporation until samples were completely dry, then resuspended in water and analysed by HPLC or LC–MS. HPLC data were collected and analysed using Chromeleon 6.8 Chromatography Data System software (Thermo Fisher).
Treatment with nuclease P1
For nuclease treatment, 100 μM of compound was incubated with 0.02 units P1 nuclease (New England Biolabs) in P1 reaction buffer (50 mM sodium acetate pH 5.5) at 37 °C for 1 h. Each reaction solution was deproteinised with a spin filter (Pall Nanosep®, MWCO 3 kDa) followed by HPLC analysis.
Liquid chromatography and mass spectrometry
Enzymatic reactions were analysed on an UltiMate 3000 UHPLC system (Thermo Fisher scientific) with absorbance monitoring at 260 nm. Samples were injected into a C18 column (Kinetex EVO 2.1 × 50 mm, particle size 2.6 µm) at 40 °C. Gradient elution was performed with solvent A (10 mM ammonium bicarbonate) and solvent B (acetonitrile plus 0.1% TFA) at a flow rate of 0.3 ml min−1 as follows: 0–0.5 min, 0% B; 0.5−3.5 min, 20% B; 3.5–5 min, 50% B; 5–7 min, 100% B.
LC–MS and LC–MS/MS analysis were carried out on a Eksigent 400 LC coupled to Sciex 6600 QTof mass spectrometer, in trap elute configuration at micro flow rates. Samples were loaded onto a YMC Triart C18 trap cartridge 0.5 ×5.0 mm in 99.95% water, 0.05% TFA at 10 µl min−1. After 3 min washing the salts to waste, the trap was switched in-line with the analytical column: a YMC Triart 150 ×0.075 mm. Gradient elution was performed with solvent A (99.9% water, 0.1% formic acid) and solvent B (20% water 80% acetonitrile 0.1% formic acid) at a flow rate of 5 µl min−1 as follows: 0 min 3% B; 0–6 min 95% B; 6–8 min 95% B; 8–9 min 3% B; 9–13 min re-equilibrate 3% B. The flow from the column sprayed directly into the ESI turbospray orifice of the mass spectrometer. Data were collected in positive ionization mode from 120–1,000 m/z. Ions of interest were selected for CID fragmentation at collision voltages of 25-45 V, and the fragmentation spectra collected from 50–1,000 m/z. The mass spectrometer was externally calibrated prior to analysis with Sciex tuning solution 4457953.
Thin-layer chromatography
Radio-labelled SAM-AMP and its analogues were separated by TLC. 1 µl radiolabelled products were analysed on 20 × 20 cm Silica gel TLC aluminium plates (Sigma-Aldrich) with 0.5 cm of TLC buffer (0.2 M ammonium bicarbonate, 70% ethanol, and 30% water pH 9.3) at 35 °C. The TLC plate was left in the TLC chamber until the solvent front was approximately 5 cm from the top of the TLC plate and finally phosphorimaged using a Typhoon FLA 7000 imager (GE Healthcare).
Electrophoretic mobility shift assay
In total, 40 nM 32P-radiolabelled SAM-AMP, SAH-AMP or cA3 was incubated with varying amounts of purified B. fragilis CorA (0.1, 0.2, 0.4, 0.8, 1.6 and 3.3 µM) in binding buffer (12.5 mM Tris-HCl, pH 8.0, 10% (v/v) glycerol, 0.5 mM EDTA) at 25 °C for 15 min. Reactions were mixed with ficoll loading buffer and then analysed on the native polyacrylamide gel (8% (w/v) 19:1 acrylamide:bis-acrylamide). Electrophoresis was carried out at 200 V for 2 h at room temperature using 1× TBE buffer as the running buffer, followed by phosphorimaging (Typhoon FLA 7000 imager (GE Healthcare), photomultiplier tube setting = 700–900).
SAM-AMP degradation assay
SAM-AMP degradation assay was carried out by incubating 1 µM wild-type B. fragilis NrN or C. botulinum SAM lyase and their inactive variants with 100 µM purified SAM-AMP, its analogues or standards like SAM, pppApA, pApA or cOA mixture (cA2, cA3, cA4 and cA6), respectively in buffer (20 mM Tris-HCl pH 7.5, 20 mM NaCl, 1% glycerol and 0.5 mM MnCl2) at 37 °C for 1 h or at indicated time points for time course assay. The reaction was stopped by mixing with 2 volumes pre-chilled methanol and vortexing for 30 s, before centrifuging at 13,000 rpm at 4 °C for 20 min to remove denatured protein. The supernatant was dried and then resuspended in the water, followed by HPLC or LC–MS/MS analysis.
Western blotting for detection of B. fragilis CorA
For detection of B. fragilis CorA wild-type and variants expression, proteins were expressed from the pEV5hisTEV plasmid in E. coli C43 (DE3). The cells were grown in 5 ml LB containing 50 µg ml−1 kanamycin at 37 °C until OD600 reached 0.6–0.8, followed by 0.2 mM IPTG induction. After growth at 16 °C for 16 h, the pellets were collected by centrifugation at 4,000 rpm for 10 min and then resuspended in 1 ml lysis buffer (50 mM HEPES, pH 7.5, 250 mM NaCl, 5% glycerol, 10 mM imidazole). Ten microlitres of 20-times-diluted lysate was loaded onto a precast NuPAGE Bis-Tris Gel (Thermo Fisher Scientific) for separation and transferred into a nitrocellulose membrane using iBlot 2 Dry Blotting System (Invitrogen). Membranes were blocked for at least 1 h with shaking in 0.03% milk in TBST (20 mM Tris, 150 mM NaCl, pH 7.6, 0.1% Tween-20) and then incubated with mouse anti-V5 antibody (Invitrogen, cat. no. R960-25, clone SV5-Pk1) diluted 1:10,000 in 0.03% milk in TBST buffer at 4 °C with overnight shaking. Membranes were washed three times with 0.03% milk in TBST and then incubated with anti-mouse IgG antibody (LI-COR Biosciences) at 1:20,000 dilution with 0.03% milk in TBST for 1 h at room temperature with shaking. The membranes were washed again with 0.03% milk in TBST twice and TBST once, before imaging on an Odyssey imager system (LI-COR Biosciences).
Nuclease assay
Nuclease activity of Cas6 was assayed by incubating 1.2 µM B. fragilis Cas6 with 300 nM 5′ end FAM-labelled B. fragilis CRISPR repeat RNA (Table S1, purchased from Integrated DNA Technologies (IDT)) in reaction buffer (20 mM Tris-HCl pH 7.5, 50 mM NaCl and 1 mM EDTA), at 37 °C for 5 min. The reaction was stopped by heating at 95 °C for 5 min and then analysed by 20% acrylamide, 7 M urea, 1× TBE denaturing gel, which was run at 30 W, 45 °C for 2 h. An alkaline hydrolysis ladder was generated by incubating RNA in 5 mM NaHCO3, pH 9.5 at 95 °C for 5 min. The gel was finally imaged using a Typhoon FLA 7000 imager (GE Healthcare) at a wavelength of 532 nm (PMT = 600–700).
An internally radiolabelled transcript RNA containing two CRISPR repeats and one guide sequence (Supplementary Table 1) was incubated with 2 µM Cas6 in the same condition mentioned above and the reaction products were checked on the 20% acrylamide denaturing gel at different time points. The transcript was generated by following the instruction of MEGAscript®Kit (Invitrogen). The template used in transcription was obtained by PCR of plasmid pCRISPR_Lpa using primer Duet-up and Duet-Down (IDT; Supplementary Table 1). PCR product (120 ng) mixed with ATP, GTP, UTP, CTP solution and 133 nM α32P-ATP as a tracer was incubated at 37 °C for 4 h in the 1× reaction buffer with T7 enzyme mix. Transcript was then purified by phenol: chloroform extraction and isopropanol precipitation.
Target RNA cleavage assay
The 5′ end labelled RNA–Lpa was generated as described previously15. RNA cleavage assays using 1 µM wild-type Cmr (or variant with Cmr4 D27A) and 5′-end-labelled RNA–Lpa substrates were conducted in reaction buffer (20 mM Tris-HCl, pH 7.5, 10 mM NaCl, 1% glycerol and 5 mM MnCl2, 0.1 U µl−1 SUPERase•In (Thermo Scientific)) at 37 °C. The reaction was stopped at indicated time points by adding EDTA (pH 8.0) and extracted with phenol–chloroform to remove protein. After adding equal volume 100% formamide, the samples were loaded onto 20% denaturing polyacrylamide sequencing gel. The gel electrophoresis was carried out at 90 W for 3–4 h. Visualization was achieved by phosphorimaging as above. A 5′-end-labelled RNA–Lpa was subjected to alkaline hydrolysis generating a single nucleotide resolution ladder for RNA size determination.
Bioinformatic analyses
To investigate the phylogenetic diversity of Cas10 proteins across type III CRISPR–Cas loci and their associations with CorA, 50,924 bacterial and 992 archaeal genomes from NCBI, marked as ‘complete’, were downloaded on 14 March 2023. The genomes were filtered for the presence of Cas10 by performing a hidden Markov model (HMM) search using Hmmer 3.3.244 and previously published Cas10 HMM profiles42. Hits with an E-value less than 10−20 and minimum protein length above 500 AA were considered. The Cas10s from the resulting 2,209 genomes were clustered with CD-hit (similarity cutoff 0.80 and word size 5) resulting in 613 representative Cas10 sequences. All CRISPR–Cas loci in the associated 613 genomes were annotated with CRISPRCasTyper 1.8.042 and type III loci (or hybrid loci with type III as a component) that had an interference completeness percentage of over 30% were picked for further analysis. This threshold was chosen to exclude any solo Cas10 proteins, but also to include small CRISPR–Cas loci that may include effector proteins such as CorA. From this point forward all CRISPR–Cas loci were treated as independent of their host, i.e multiple type III loci from a single host were permitted, resulting in 745 loci. Proteins within these loci (and 2 kb upstream and downstream of the CCTyper-reported locus boundaries) were searched for CorA using Hmmer 3.3.244 with HMM profiles derived from CCTyper42 using an E-value cutoff of 10−20. To create the Cas10 phylogenetic tree, the Cas10 proteins from each locus were aligned using Muscle 5.145 with the Super5 algorithm intended for large datasets. The Cas10 phylogenetic tree was constructed from the alignment using FastTree 2.1.1146 with the WAG + CA model and Gamma20-based likelihood. The tree was visualized in R 4.1.1 and RStudio 2021.9.0.351 (http://www.rstudio.com/) using ggtree47 and ggplot248. These steps were wrapped in a Snakemake 7.22.049 pipeline and an R script (available at Github: https://github.com/vihoikka/Cas10_prober).
Structural modelling of B. fragilis Cas10 and B. fragilis CorA
The structure of the B. fragilis Cas10–Cas5 heterodimer and B. fragilis CorA monomer were predicted using AlphaFold250,51 (AF2) as implemented by the Colabfold server52. The structure of the C-terminal 172 amino acids of CorA, including the transmembrane domain, which is the region with detectable similarity to the CorA magnesium transporter, was modelled as a pentamer using Alphafold2. Five individual monomers were then superimposed on the pentameric transmembrane structure to yield a model of the full-length pentameric B. fragilis CorA structure. This approach avoided memory and processor constraints inherent in Colabfold for such a large modelling project. The local distance different test (LDDT) scores for all three models are shown in Extended Data Fig. 10.
Statistics and reproducibility
The CorA binding assay in Fig. 4a was repeated as duplicates. Assays for nuclease activity of Cas6 (Extended Data Fig. 2) and target RNA cleavage activity of B. fragilis Cmr complex (Extended Data Fig. 3) were conducted as three independent experiments and representative examples are shown. The purification of CorA (Extended Data Fig. 8a) was repeated three times and the western blot of CorA (Extended Data Fig. 8e) was performed as two independent experiments (two biological replicates) with comparable results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Mass spectrometry data are available on FigShare: https://doi.org/10.6084/m9.figshare.c.6646859.v1. Coordinates for experimentally determined protein structures (for example Protein Data Bank ID 3W2W) are available at https://www.rcsb.org/. The genome sequences used to make Fig. 1a were downloaded from NCBI (https://www.ncbi.nlm.nih.gov) on 14 March 2023.
Code availability
Code used to generate the phylogenetic tree was wrapped in a Snakemake 7.22.049 pipeline and an R script that are available in Github: https://github.com/vihoikka/Cas10_prober.
References
-
Makarova, K. S. et al. An updated evolutionary classification of CRISPR–Cas systems. Nature Rev. Microbiol 13, 722–736 (2015).
-
Makarova, K. S. et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 18, 67–83 (2020).
-
Samai, P. et al. Co-transcriptional DNA and RNA cleavage during type III CRISPR–Cas immunity. Cell 161, 1164–1174 (2015).
-
Elmore, J. R. et al. Bipartite recognition of target RNAs activates DNA cleavage by the Type III-B CRISPR–Cas system. Genes Dev. 30, 447–459 (2016).
-
Kazlauskiene, M., Tamulaitis, G., Kostiuk, G., Venclovas, C. & Siksnys, V. Spatiotemporal control of type III-A CRISPR–Cas immunity: coupling DNA degradation with the target RNA recognition. Mol. Cell 62, 295–306 (2016).
-
Kazlauskiene, M., Kostiuk, G., Venclovas, C., Tamulaitis, G. & Siksnys, V. A cyclic oligonucleotide signaling pathway in type III CRISPR–Cas systems. Science 357, 605–609 (2017).
-
Niewoehner, O. et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017).
-
Rouillon, C., Athukoralage, J. S., Graham, S., Gruschow, S. & White, M. F. Control of cyclic oligoadenylate synthesis in a type III CRISPR system. eLife 7, e36734 (2018).
-
Steens, J. A., Salazar, C. R. P. & Staals, R. H. J. The diverse arsenal of type III CRISPR–Cas-associated CARF and SAVED effectors. Biochem. Soc. Trans. 50, 1353–1364 (2022).
-
Athukoralage, J. S. & White, M. F. Cyclic oligoadenylate signalling and regulation by ring nucleases during type III CRISPR defence. RNA 27, 855–867 (2021).
-
Wexler, A. G. & Goodman, A. L. An insider’s perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2, 17026 (2017).
-
Shkoporov, A. N. et al. PhiCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun. 9, 4781 (2018).
-
Wexler, H. M. Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007).
-
Tajkarimi, M. & Wexler, H. M. CRISPR–Cas systems in Bacteroides fragilis, an important pathobiont in the human gut microbiome. Front. Microbiol. 8, 2234 (2017).
-
Grüschow, S., Adamson, C. S. & White, M. F. Specificity and sensitivity of an RNA targeting type III CRISPR complex coupled with a NucC endonuclease effector. Nucleic Acids Res, 49, 13122–13134 (2021).
-
Shmakov, S. A., Makarova, K. S., Wolf, Y. I., Severinov, K. V. & Koonin, E. V. Systematic prediction of genes functionally linked to CRISPR–Cas systems by gene neighborhood analysis. Proc. Natl Acad. Sci. USA 115, E5307–E5316 (2018).
-
Shah, S. A. et al. Comprehensive search for accessory proteins encoded with archaeal and bacterial type III CRISPR–Cas gene cassettes reveals 39 new cas gene families. RNA Biol. 16, 530–542 (2019).
-
Heo, K. et al. A pGpG-specific phosphodiesterase regulates cyclic di-GMP signaling in Vibrio cholerae. J. Biol. Chem. 298, 101626 (2022).
-
Wang, F. et al. Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase. Biochem. J. 475, 191–205 (2018).
-
Hsiao, Y. Y., Duh, Y., Chen, Y. P., Wang, Y. T. & Yuan, H. S. How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes. Nucleic Acids Res. 40, 8144–8154 (2012).
-
Lee, C. W. et al. Structural basis of small RNA hydrolysis by oligoribonuclease (CpsORN) from Colwellia psychrerythraea strain 34H. Sci. Rep. 9, 2649 (2019).
-
Nicholls, T. J. et al. Dinucleotide degradation by REXO2 maintains promoter specificity in mammalian mitochondria. Mol. Cell 76, 784–796.e786 (2019).
-
Guo, X. et al. Structure and mechanism of a phage-encoded SAM lyase revises catalytic function of enzyme family. eLife 10, e61818 (2021).
-
Simon-Baram, H. et al. SAMase of bacteriophage T3 inactivates Escherichia coli’s methionine S-adenosyltransferase by forming heteropolymers. mBio 12, e0124221 (2021).
-
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
-
Grüschow, S., Athukoralage, J. S., Graham, S., Hoogeboom, T. & White, M. F. Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucl. Acids Res. 47, 9259–9270 (2019).
-
Shao, Y. et al. A non-stem-loop CRISPR RNA is processed by dual binding Cas6. Structure 24, 547–554 (2016).
-
Tamulaitis, G., Venclovas, C. & Siksnys, V. Type III CRISPR–Cas immunity: major differences brushed aside. Trends Microbiol. 25, 49–61 (2017).
-
Tamulaitis, G. et al. Programmable RNA shredding by the type III-A CRISPR–Cas system of Streptococcus thermophilus. Mol. Cell 56, 506–517 (2014).
-
Estrella, M. A., Kuo, F. T. & Bailey, S. RNA-activated DNA cleavage by the type III-B CRISPR–Cas effector complex. Genes Dev. 30, 460–470 (2016).
-
Vedel, M., Lawrence, F., Robert-Gero, M. & Lederer, E. The antifungal antibiotic sinefungin as a very active inhibitor of methyltransferases and of the transformation of chick embryo fibroblasts by Rous sarcoma virus. Biochem. Biophys. Res. Commun. 85, 371–376 (1978).
-
Halliday, N. M., Hardie, K. R., Williams, P., Winzer, K. & Barrett, D. A. Quantitative liquid chromatography–tandem mass spectrometry profiling of activated methyl cycle metabolites involved in LuxS-dependent quorum sensing in Escherichia coli. Anal. Biochem. 403, 20–29 (2010).
-
Osawa, T., Inanaga, H. & Numata, T. Crystal structure of the Cmr2–Cmr3 subcomplex in the CRISPR–Cas RNA silencing effector complex. J. Mol. Biol. 425, 3811–3823 (2013).
-
Jia, N., Jones, R., Sukenick, G. & Patel, D. J. Second messenger cA4 formation within the composite Csm1 palm pocket of type III-A CRISPR–Cas Csm complex and its release path. Mol. Cell 75, 933–943 e936 (2019).
-
VanderWal, A. R. et al. Csx28 is a membrane pore that enhances CRISPR–Cas13b-dependent antiphage defense. Science 380, 410–415 (2023).
-
Tal, N. et al. Cyclic CMP and cyclic UMP mediate bacterial immunity against phages. Cell 184, 5728–5739.e5716 (2021).
-
Whiteley, A. T. et al. Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature 567, 194–199 (2019).
-
Duncan-Lowey, B., McNamara-Bordewick, N. K., Tal, N., Sorek, R. & Kranzusch, P. J. Effector-mediated membrane disruption controls cell death in CBASS antiphage defense. Mol. Cell 81, 5039–5051.e5035 (2021).
-
Athukoralage, J. S. et al. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature 577, 572–575 (2020).
-
Velisetty, P. & Chakrapani, S. Desensitization mechanism in prokaryotic ligand-gated ion channel. J. Biol. Chem. 287, 18467–18477 (2012).
-
Zhang, J. & Zheng, Y. G. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol. 11, 583–597 (2016).
-
Russel, J., Pinilla-Redondo, R., Mayo-Munoz, D., Shah, S. A. & Sorensen, S. J. CRISPRCasTyper: automated identification, annotation, and classification of CRISPR–Cas loci. CRISPR J 3, 462–469 (2020).
-
Rouillon, C., Athukoralage, J. S., Graham, S., Grüschow, S. & White, M. F. Investigation of the cyclic oligoadenylate signalling pathway of type III CRISPR systems. Methods Enzymol. 616, 191–218 (2019).
-
Eddy, S. R. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 (2011).
-
Edgar, R. C. Muscle5: high-accuracy alignment ensembles enable unbiased assessments of sequence homology and phylogeny. Nat. Commun. 13, 6968 (2022).
-
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
-
Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinformatics 69, e96 (2020).
-
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009).
-
Molder, F. et al. Sustainable data analysis with Snakemake. F1000Research 10, 33 (2021).
-
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
-
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucl. Acids Res. 50, D439–D444 (2022).
-
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Acknowledgements
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (Grant BB/T004789/1 to M.F.W.) and European Research Council (ref. 101018608 to M.F.W.). H.C. acknowledges the support of the China Scholarship Council (code 202008420207). V.H. is funded by the Finnish Cultural Foundation. The authors thank D. O’Hagan and R. White for helpful discussions. For the purpose of open access, the author has applied a CC BY public copyright Licence to any Accepted Author Manuscript version arising from this submission.
Author information
Authors and Affiliations
Contributions
H.C. planned, carried out and analysed the molecular biology, microbiology and biochemistry experiments and drafted the manuscript. V.H. carried out the bioinformatic analyses. S. Grüschow planned the protein expression and plasmid challenge experiments with H.C. S.Graham cloned genes and expressed proteins. S.S. carried out the mass spectrometry and analysed the data. M.F.W. conceptualized and oversaw the project, obtained funding and analysed data along with the other authors. All authors contributed to the drafting and revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Eugene Koonin, Philip Kranzusch and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Plasmid challenge assay.
a, Plasmid pBfrCmr1-6 was constructed by Gibson assembly to express genes cmr1–cmr6 (junctions indicated by black squares); Plasmid pCRISPR was constructed to express the CRISPR array along with Cas6. Plasmid maps were generated by Benchling (https://benchling.com/). b, Different variants of ancillary effectors were tested in the B. fragilis Cmr wild type target (tetR) and non-target (pUC19) system. CorAtr is the truncated CorA (aa 1-428) and NrNΔ is an inactive variant of NrN (D85A/H86A/H87A). The schematic shows cartoons of the three plasmids used in these experiments. c, The ancillary effectors CorA and NrN were assayed in the Mycobacterium tuberculosis (Mtb) Cmr wild type system. TsuCsx1 from Thioalkalivibrio sulfidiphilus (activated by cA4) acted as a positive control. Uncropped images and gels are available in Supplementary Data Fig. 4.
Extended Data Fig. 2 Expression of the B. fragilis Cmr complex and CRISPR repeat RNA processed by Cas6.
a, SDS-PAGE analysis of the purified wild type (WT) and variants of Cmr (1-6) complex which include Cmr2 D328A:D329A (∆cy), Cmr2 D70N (DN), Cmr2 E151R (ER) and the double mutant (D70N:E151R, DNER). b, The cleavage site of Cas6 within the CRISPR repeat RNA was mapped by incubating 5′ end FAM-labelled repeat RNA (300 nM) with Cas6 nuclease (1.2 µM). Alkaline hydrolysis (OH−) ladder was used to mark the size of 5′ RNA cleavage products (green arrow). Potential secondary structure of CRISPR repeat RNA with cleavage site was indicated (green arrow). c, An internally radio-labelled transcript RNA containing two CRISPR repeats (blue) and one guide (targeting Phage P1) sequence (orange) was incubated with BfrCas6 (2 µM). Samples were collected at the indicated time points and analysed by denaturing gel. The expected sizes and compositions of cleavage products are indicated based on the specific cleavage site of Cas6 within each repeat. Uncropped gels are available in Supplementary Data Figs. 3–5.
Extended Data Fig. 3 B. fragilis Cmr crRNA composition and target RNA degradation.
a, The sequence of crRNA species extracted from purified Cmr and the target RNA substrate used in the activity assay. The repeat-derived sequence (8 nt tag), spacer-derived sequence (guide) and the sequence complementary to guide RNA are coloured black, red and green, respectively. Five putative cleavage sites are indicated by arrows (Site1 is indicated by purple arrows, while sites 2 to 5 by red arrows). Extracted crRNAs and the target RNA substrate were 5′-labelled with 32 P (blue star). b, The size of extracted crRNAs from wild type and mutant Cmr (Cmr4 D27A) was mapped by comparing with alkaline hydrolysis ladder of Target_LPA substrates (L1-3 with increased concentration of substrates). c, The indicated Target_LPA was incubated with (+) or without (−/C1) wild type Cmr in the presence of Mn2+(no Mn2+ in buffer C2). The cleavage sites were mapped by comparing with alkaline hydrolysis (OH-) ladder and indicated by red arrows. d, Time course of cleavage on the 5′-radio-labelled Target_LPA by wild type or mutant Cmr (Cmr4 D27A). The buffer of C1 and C2 are in absence of Mn2+, while C0 is in absence of Cmr. Uncropped gels are available in Supplementary Data Fig. 5.
Extended Data Fig. 4 In vitro characterisation of B. fragilis Cmr-catalysed reaction.
a, ATP (0.5 mM) and SAM (0.5 mM) were incubated with wild type of Cmr (3 µM) in presence of Mn2+. Samples were collected at the indicated time points and analysed by HPLC. Cmr was omitted in control samples. b, as for a, with SAH in place of SAM. c, HPLC and MS analysis of Cmr reaction products SAM-AMP, SAH-AMP and sinefungin-AMP. Uncropped HPLC traces are available in Supplementary Data Fig. 6.
Extended Data Fig. 5 P1 nuclease degradation of reaction products.
HPLC analysis of nuclease P1-mediated hydrolysis reactions towards Cmr’s products (left). cA2 (cyclic di-3′,5′-adenylate) and pApA (5′- phosphoadenylyl- (3′ → 5′)- adenosine) were used as controls. The proposed reactions of P1 were shown on the right. Uncropped HPLC traces are available in Supplementary Data Fig. 6.
Extended Data Fig. 6 Alignment of Cas10 sequences from the CorA-1 cluster.
Key binding site residues shown in Fig. 3 are indicated. Sequence IDs are: Bacteroides fragilis ANQ60746.1; Clostridium botulinum WP_011986674; Prevotella - Xylanibacter muris WP_172276208; Camphylobacterales bacterium HIP52383.1; Aliarcobacter butzleri WP_260918755; Syntrophothermus lipocalidus WP_013175521; Methanococcus voltae WP_209731901).
Extended Data Fig. 7 pppApA synthesis by the wild-type and variant BfrCmr complexes.
In vitro pppApA synthase activity of wt and variants B. fragilis Cmr, analysed by HPLC following incubation of 2 µM Cmr with 0.5 mM ATP for 30 min. The D70N/E151R double mutant synthesises pppApA but not SAM-AMP. Uncropped HPLC traces are available in Supplementary Data Fig. 7.
Extended Data Fig. 8 Purification, modelling and mutagenesis of the CorA protein.
a, SDS-PAGE analysis of the final gel filtration step for purification of B. fragilis CorA, which was used for SAM-AMP binding assays. The four tightly spaced protein bands in the gel all correspond to CorA, perhaps indicating limited proteolysis of the termini. M – M.Wt. markers, S – sample applied to gel filtration; the horizontal bar indicates the three fractions pooled for further analysis. b, Top-down view of the pentameric BfrCorA model with individual subunits coloured differently and the conserved R152/R152/D219/D220 residues indicated by black spheres. c, Orthogonal view of the BfrCorA model showing the TM helical bundle at the bottom. d, Close up of the intersubunit interface for CorA, with conserved residues (defined in Supplementary Data Fig. 2) shown and a model of SAM-AMP in sphere representation included for scale. e, Western blot using the V5 antibody to detect expression of the wild-type (WT), R152A/R153A (RA) and D219A/D220A (DA) variants in E. coli. f, Plasmid challenge assay, showing that wild-type CorA in conjunction with NrN provides immunity from plasmids carrying a target sequence, but neither CorA variant does. Uncropped images and gels are available in Supplementary Data Fig. 8.
Extended Data Fig. 9 Specificity of the NrN and SAM-AMP lyase effectors.
a, HPLC analysis of reaction products when the NrN PDE was incubated with cA2, cA3, cA4, cA6, pApA and SAM-AMP for 30 min. Only SAM-AMP is a substrate for NrN. b, HPLC analysis of reaction products for SAM-AMP and SAM following incubation with C. botulinum SAM-AMP lyase for 30 min. SAM-AMP is preferred as a substrate over SAM. Uncropped gel and HPLC traces are available in Supplementary Data Fig. 9.
Extended Data Fig. 10 Local distance difference test (IDDT) scores for Alphafold2 models.
a, BfrCas10−5 complex; b, pentameric BfrCorA C-terminal TM domain; c, monomeric full-length BfrCorA protein.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-10 and Supplementary Table 1
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chi, H., Hoikkala, V., Grüschow, S. et al. Antiviral type III CRISPR signalling via conjugation of ATP and SAM. Nature (2023). https://ift.tt/9dpEhSV
-
Received:
-
Accepted:
-
Published:
-
DOI: https://ift.tt/9dpEhSV
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
"type" - Google News
October 18, 2023 at 10:11PM
https://ift.tt/6rBOu8W
Antiviral type III CRISPR signalling via conjugation of ATP and SAM - Nature.com
"type" - Google News
https://ift.tt/J1zelvs
https://ift.tt/TFpOS9E
Bagikan Berita Ini














0 Response to "Antiviral type III CRISPR signalling via conjugation of ATP and SAM - Nature.com"
Post a Comment