The characteristic shapes and functions of proteins depend on the order in which the constituent amino acids are linked into chains. However, further chemical modifications are often made after the amino acids are strung together. These modifications include crosslinks between certain amino-acid residues. The most common type of crosslink is a disulfide: two sulfur atoms connected by a covalent bond. Writing in Nature, Wensien et al.1 report an entirely different type of protein crosslink, in which an oxygen atom connects a nitrogen to a sulfur atom. Moreover, the authors present evidence that such N–O–S bridges have gone unnoticed in previously reported structural analyses of other proteins.
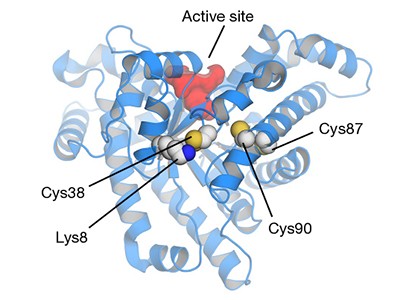
Wensien and colleagues were studying the enzyme transaldolase from Neisseria gonorrhoeae, a bacterium that causes the eponymous sexually transmitted disease. They observed that the purified enzyme was almost inactive, but that activity could be restored by using reducing agents commonly used to break disulfide bonds.
To form a disulfide, the side chains of two cysteine amino-acid residues — which can be far apart along the protein chain — must come close together in space. Noticing that the transaldolase contains a few cysteine residues, the authors reasoned that two of them might form a disulfide that inactivates the enzyme. However, when they replaced each of the cysteines individually with another amino acid, they found that only one of the resulting mutant enzymes resisted inactivation. If the enzyme really did contain an inactivating disulfide, then replacement of either of the two participating cysteines would be expected to have this effect (although there are exceptions to this simple generalization2). So what was going on?
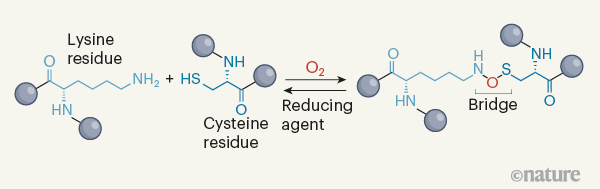
Wensien et al. solved the puzzle by using X-ray crystallography to determine the structure of the transaldolase at atomic resolution. This analysis revealed a covalent connection between a cysteine and a lysine residue — an N–O–S bridge — instead of a cysteine–cysteine disulfide crosslink (Fig. 1). Intriguingly, the oxygen atom in the N–O–S bridge does not arise from the groups in the side chains of either of those residues (the cysteine side chain contains a thiol (SH) group, whereas the lysine side chain contains an amine (NH2) group). However, the authors saw an oxygen molecule close to these side chains in the crystal structure of the reduced form of the protein (which lacks the crosslink). This observation supports the authors’ reasonable speculation that an oxygen molecule contributes an oxygen atom to the N–O–S bridge.
The discovery of an N–O–S bridge in proteins is noteworthy because non-biological reactions that produce such a molecular motif are not known, with the possible exception of one unusual molecule (a type of cyclic aromatic compound3). The formation of an N–O bond in small-molecule chemistry requires strong oxidizing conditions4, but such conditions would probably also convert sulfur atoms to higher oxidation states than that of the sulfur in the N–O–S bridge. Furthermore, small molecules containing the N–O–S motif might be in danger of undergoing disproportionation — a process in which two of the same molecules react with one another to produce two different products. In the context of a protein, favourable positioning of the cysteine’s thiol, the lysine’s amine and an oxygen molecule might aid the oxidation needed for N–O–S formation, whereas spatial (steric) constraints imposed by the surrounding protein structure might stabilize the crosslink and block further oxidation of the sulfur.
The authors speculate about possible mechanisms for bridge formation, favouring a reaction in which hydroxyl groups (OH) are added to both the sulfur atom of the cysteine and the amine group of the lysine side chain (see Extended Data Fig. 3b of ref. 1). Given the novelty of the N–O–S crosslink and the focus of the study on structural biology, important details of the chemical mechanism remain to be addressed. Specifically, the way in which the oxygen molecule is activated to take part in this reaction is not described. Many chemical reactions with molecular oxygen involve free radicals5, and so radical pathways should be explicitly considered in mechanisms for generating the N–O–S bridge.
Another mechanistic issue is how the crosslink affects enzymatic activity. The structure of the catalytic site of the crosslinked transaldolase differs only slightly from that of the non-crosslinked version of the enzyme. How formation of the N–O–S bridge inhibits catalysis is thus not obvious. The authors focused their attention on these minor structural differences, but they also observed that the crosslinked transaldolase is more resistant to heat-induced unfolding than is the non-crosslinked version. This result is not surprising and implies that the crosslinked enzyme undergoes fewer conformational fluctuations that could lead to unfolding. Smaller-scale fluctuations could be required for catalytic activity, and might also be restrained by the presence of the crosslink.
There is a key conceptual difference between the use of disulfide bonds for regulating protein function and the use of an N–O–S bridge. The formation of disulfide bonds is chemically reversible, which means that, in biological systems, disulfides are frequently made and broken in ‘exchange’ reactions with other molecules that contain disulfides or thiols. By contrast, the N–O–S linkage is formed by a different chemistry from that by which it is cleaved — that is, molecular oxygen is used to form the bridge but is not released when the bridge is broken.
Moreover, the thermodynamics of these reactions indicate that it is difficult to form the N–O–S bridge but easy to break it. N–O–S crosslinks therefore might have evolved to enable the selective activation of an enzyme under conditions in which disulfides are preserved. The particular advantages conferred by the N–O–S bridge, and the biological scenarios in which it is more useful than a disulfide, can now be explored.
The discovery of a new protein linkage has implications beyond the specifics of the enzyme studied and of the N–O–S crosslink itself. Perhaps surprisingly, the task of generating structural models for proteins is sometimes more difficult when high-resolution X-ray data are available. Variations in protein conformation or in chemical composition might be buried in the noise of the data at low resolution, but this heterogeneity becomes visible at high resolution and must therefore be interpreted6. Unanticipated chemical groups might also be lurking in the data. Wensien and colleagues’ study will inspire structural biologists to investigate deviations from expectation in their electron-density maps of biomolecules.
It has long been appreciated that enzymes are the world’s best organic chemists, because they can promote reactions that would be almost impossible in their absence. The findings show that the covalent chemistry of enzymes themselves can also defy chemical intuition.
"type" - Google News
May 05, 2021 at 10:23PM
https://ift.tt/3elknoL
Previously unknown type of protein crosslink discovered - Nature.com
"type" - Google News
https://ift.tt/2WhN8Zg
https://ift.tt/2YrjQdq
Bagikan Berita Ini














0 Response to "Previously unknown type of protein crosslink discovered - Nature.com"
Post a Comment